Project 2
Structure-Based Drug Discovery
Subproject 4: Drug Discovery and Design
SPID: 0003
Structural and Biological Identification of Residues on the Surface of NS3 Helicase
Required for Optimal Replication of the Hepatitis C Virus
Kevin Raney, Joshua Sakon
Background: Hepatitis C virus (HCV) infects over 170 million persons worldwide. It is the leading cause of liver disease in the United States and is responsible for most liver transplants. Current treatments for this infectious disease are inadequate; therefore, new therapies must be developed. HCV replication is believed to require a multiprotein complex involving NS3 helicase proteins, membrane components, and other cellular proteins. Previous studies suggest that multiple copies of NS3 helicase are utilized for the replication of a single RNA genome of HCV.
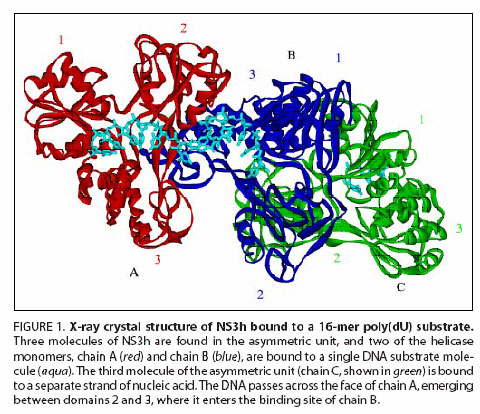
Advance: Researchers at the University of Arkansas solved the X-ray crystal structure of a complex in which a 16mer oligonucleotide was bound to two molecules of NS3 helicase (Figure). Several amino acid residues at the interface of the two NS3h molecules were identified that appear to mediate protein-protein interactions. Mutations introduced to disrupt this interface wee found to result in a dramatic reduction in replication capacity. However, the biochemical activities of the mutant enzyme do not reflect the large reduction in HCV replicative capacity revealed in the biological experiments. Hence, the known, measurable biochemical properties associated with the helicase activity of NS3h do not reveal all of the likely biological roles of the helicase domain.
Implications: Current treatments for Hepatitis C virus are inadequate, and therefore it is important to develop new strategies to treat this disease. It is believed that a better understanding of the structure and mechanism of the NS3 helicase that carries out replication of HCV will lead to the development of a new class of novel helicase inhibitors targeting the Hepatitis C virus
Publications
- EOFF, R. E., SPURLING, T. L., AND RANEY, K. D. “Chemically modified DNA substrates
implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase”
Biochemistry 44, 666-674. 2005
Acknowledges COBRE - TACKETT, A. J., CHEN, Y., CAMERON, C. E., AND RANEY, K. D. “Multiple Full-length NS3
Molecules are Required for Optimal Unwinding of Oligonucleotide DNA in vitro”, J.
Biol. Chem., 280, 10797-10806. 2005
Acknowledges COBRE - MACKINTOSH, S. G., LU, Z. L., JORDAN, J. B., HARRISON, M. K., SIKORA, B., SHARMA,
S. D., CAMERON, C. E., AND RANEY, K. D.* AND SAKON, J.* “Identification of residues
on the surface of NS3 helicase that are required for optimal replication of the Hepatitis
C Virus” J. Biol. Chem. 281, 3528-3535, 2006.
Acknowledges COBRE - EOFF, R. L., AND RANEY, K. D. “Intermediates revealed in the kinetic mechanism for
DNA unwinding by a monomeric helicase” in press, Nat. Struct. and Mol. Biol. (2006)
Acknowledges COBRE - SPURLING, T. L., EOFF, R. L., AND RANEY, K. D. “Dda Helicase Unwinds a DNA-PNA Chimeric
Substrate: Evidence for an Inchworm Mechanism” in press, Bioorganic and Med. Chem.
Letters. (2006)
Acknowledges COBRE