Project 3
Protein Targeting
Subproject 3: Protein Targeting
SPID: 0004
Robyn Goforth, Suresh Kumar, Ralph Henry, Chin Yu, Joshua Sakon
Signal recognition particle (SRP) and its receptor (SRa/SRß) are GTPases that are used to target newly synthesized polypeptides to a protein conducting transporter in the endoplasmic reticulum. Malfunction of the SRP targeting/transport system in humans is implicated in several human diseases including idiopathic inflammatory myopathies and diabetes mellitus. We are using a model SRP in chloroplasts (cpSRP) to develop a structure-based understanding of the SRP targeting mechanism. Unlike mammalian and bacterial SRPs, cpSRP uniquely functions in the absence of behemoth ribosomes and cpSRP interaction with targeting substrates (e.g. LHCP) can be reconstituted in vitro (See Fig. B). Hence, it is possible to combine structural and biochemical methods to gather a detailed understanding of the SRP targeting mechanism.
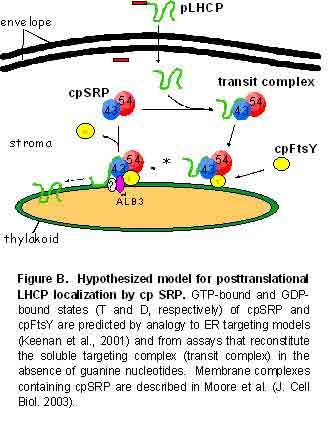
cpSRP is a heterodimeric stromal protein composed of a SRP54 homologue (cpSRP54) and a novel 43 kDa subunit (cpSRP43). cpSRP43 is unique to chloroplasts and is dominated by predicted protein interaction motifs that include three chromatin-binding domains (CD1, CD2 and CD3) and four ankyrin (Ank)-binding domains. Recent studies with chromo domain deletion constructs reveal that each chromo domain of cpSRP43 has a distinctly different role in the post-translational localization of thylakoid proteins. CD2 is found to be essential for both targeting complex formation and integration. CD2 is also demonstrated to contribute to the binding of cpSRP54. CD1 is shown to be not necessary for targeting complex formation but it is absolutely essential for integration. The role of the C-terminal chromo domain (CD3) is not still clear. Characterization of the structural features of the individual chromo domains will provide the necessary insights required to comprehend their observed functional differences.
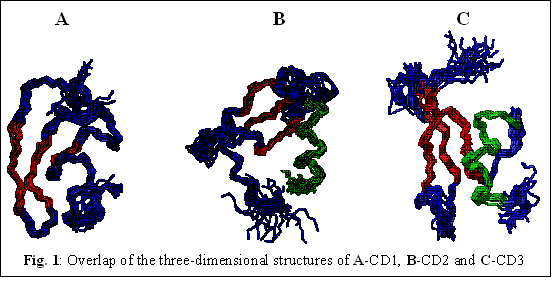
Three-dimensional solution structures of the CD domains of cpSRP43: We have successfully solved the three-dimensional solution structures of all the three chromatin domains present in cpSRP43 using multidimensional NMR techniques. The secondary structural elements in CD2 and CD3 include a triple-stranded beta-sheet and a c-terminal ?-helix (Figs. 1B and 1C). Interestingly, the orientation of the ?-helix in CD2 and CD3 are strikingly different. In CD2, the helical segment is positioned perpendicular to the plane of the beta-sheet. In CD3, the c-terminal helix is aligned parallel to the triple-stranded beta-sheet (Fig. 1C). The three-dimensional structure of CD1 consists of three beta strands arranged to form a beta-barrel type of structure (Fig. 1A). CD1 lacks the helical segment found in both CD2 and CD3. Although the structures of CD1, CD2 and CD3 are grossly similar, the chromatin domains of cpSRP43 differ significantly in their surface charge potentials.
Binding sites of cpSRP54 on CD2: Isothermal titration calorimetry experiments show that cpSRP54 binds only to CD2 of cpSRP43. Therefore, the cpSRP54 binding sites on CD2 were mapped using 1H-15N chemical shift perturbation monitored by 1H-15N HSQC spectra. It is observed that binding of cpSRP54 does not cause any significant conformational changes in CD2. The cpSRP54 binding sites in CD2 are mostly distributed in the c-terminal helix and in beta strand -1. Val10, Asp11, Glu15 and Arg17 in beta strand-1 and Val47, Lys49, Asp53 and Leu55 (in the c-terminal helix) constitute the cpSRP54 binding surface.
Publications
- Cai, Y.; Moore, Misty; Goforth, Robyn L.; Henry, Ralph; Beitle, Robert. Genomic data for alternate production strategies: Identification of major contaminating species for Co(II) immobilized metal affinity chromatography. Biotechnology and Bioengineering 88 (1), 77-83 (2004) NIH NCRR COBRE Grant 1 P20 RR15569 acknowledged
- Goforth, Robyn L.; Peterson, Eric C.; Yuan, Jianguo; Moore, Misty; Kight, Alicia; Lohse, Matthew B.; Sakon, Joshua; Henry, Ralph L. Regulation of the GTPase cycle in posttranslational signal recognition particle based protein targeting involves cpSRP43. J. Biol. Chem. 279, 43077-84 (2004) NIH NCRR COBRE Grant 1 P20 RR15569 acknowledged