Project 4
Principles of Protein Folding and Design
Subproject 1: Structural Characterization of the Fibroblast Growth Factor Signaling
Complex
SPID: 0005
Suresh Kumar
 Fibroblast growth factors are potent mitogens that regulate key cellular processes
such as angiogenesis, tumor growth, wound healing and differentiation. FGFs exhibit
their biological activity by binding to their high affinity cell surface receptor,
FGFR. Aberrant activation of the FGF signaling pathway is responsible for the onset
of tumors and several craniosynostosis and dwarfing syndromes, such as Apert syndrome,
Pfeiffer syndrome, and achondroplasia. The prototype FGFR consists of an extracellular
ligand binding domain that contains three immunoglobulin-like domains (D1, D2 and
D3), a transmembrane domain and a cytoplasmic domain that bears the tyrosine kinase
activity. FGF signaling is triggered by binding of the ligand (FGF) to the extracellular
domain of FGFR. The D2, D3 modules of the extracellular ligand binding domain are
well conserved and are shown to be minimal unit sufficient for the specific ligand
(FGF) binding activity.
Fibroblast growth factors are potent mitogens that regulate key cellular processes
such as angiogenesis, tumor growth, wound healing and differentiation. FGFs exhibit
their biological activity by binding to their high affinity cell surface receptor,
FGFR. Aberrant activation of the FGF signaling pathway is responsible for the onset
of tumors and several craniosynostosis and dwarfing syndromes, such as Apert syndrome,
Pfeiffer syndrome, and achondroplasia. The prototype FGFR consists of an extracellular
ligand binding domain that contains three immunoglobulin-like domains (D1, D2 and
D3), a transmembrane domain and a cytoplasmic domain that bears the tyrosine kinase
activity. FGF signaling is triggered by binding of the ligand (FGF) to the extracellular
domain of FGFR. The D2, D3 modules of the extracellular ligand binding domain are
well conserved and are shown to be minimal unit sufficient for the specific ligand
(FGF) binding activity.
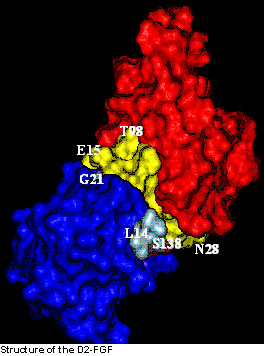
The three-dimensional solution structure of the D2 domain of FGFR was determined (at 0.4Å) using a variety of triple resonance NMR techniques. The binding interface of the D2 domain – FGF binary complex has been characterized using 1H-15N chemical shift perturbation, residual dipolar coupling measurements and amide proton exchange experiments. The residues in FGF that participate in the receptor binding are mostly located in beta-strands II, and IX, and the loop between beta-strands XI and XII. The residues in the D2 domain that bind to FGF are Glu15, Met17, Glu18, Arg20, His22-Asn28.
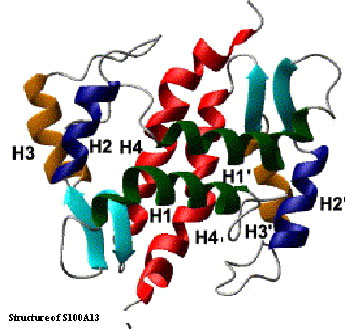
FGFs lack the N-terminal signal sequence required for its release through the endoplasmic recticulum – Golgi apparatus secretory pathway. However, FGF’s are required to be released into the extracellular compartment to exhibit their biological functions by binding to their cell surface receptor. Recent studies have shown that FGF is secreted into the extracellular compartment as a multiprotein complex consisting of FGF, S100A13 (a calcium binding protein) and synaptotagmin (a calcium sensor in neurotransmitter release). Copper is shown to play an important role in the formation/organization of the multiprotein release complex. As a first step towards our goal of understanding the three-dimensional structure of the multiprotein FGF release complex, we have determined the three-dimensional solution structure of S100A13 at high resolution using a wide range of multidimensional NMR techniques. S100A13 is a homodimer consisting of four helices. There are two helix-loop-helix EF-hands in each monomeric unit of S100A13. Two calcium ions bind per monomer of the protein. Isothermal titration calorimetry studies show that S100A13 binds to copper ions with reasonably high affinity (Kd ~ 10-7 M). 1H-15N chemical shift perturbation experiments reveal the presence of a unique copper binding site in the structure of S100A13. Our data suggest that the copper bound to S100A13 is responsible for the oxidation of the thiol group of cysteine 30 in FGF.
Significance: Characterization of the FGF-receptor interactions and understanding the nature of
interactions that are involved in the formation and stabilization of the multiprotein
FGF release complex are expected to provide useful clues for the rational design of
therapeutic principles against a variety of FGF-mediated diseases. In addition, determination
of the structures of proteins and protein complexes involved in the cpSRP pathway
will provide valuable insights into the mechanism underlying the targeting of important
plant proteins such as the light harvesting chlorophyll binding protein.
Publications
- KATHIR, KARUPPANAN M.; KUMAR, SURESH T. K.;.YU, CHIN. Understanding the Anti-Mitogenic Activity of Suramin Biochemistry 45(3):899-906. 2006 (COBRE funding acknowledged).
- SIVARAJA, VAITHYALINGAM.; KUMAR, SURESH T. K.; LEENA, PHILOMINATHAN S. T.; CHANG, AN-NI; VIDYA, CHITOORI; GOFORTH, ROBYN.;. RAJALINGAM, DAKSHINAMURTHY.; KANNAN, ARVIND.; YE, JIANG, LIANG; CHOU, JONATHAN.; HENRY, RALPH.; YU, CHIN. Three-dimensional Solution Structures of the Chromo Domains of cpSRP43. J. Biol. Chem. 280(50):41465-71 2005 (COBRE funding acknowledged)
- HUNG, KUO-WEI,; KUMAR, SURESH T.K.; KATHIR, KARUPANAN.; XU,PING.; NI, FENG.; JI, HSIANG-HUI.; CHEN, MEI-CHI.; YANG, CHU-CHI.; LIN, FU-PANG.; YU, CHIN. Solution Structure of the Ligand-Binding Domain of the Fibroblast Growth Factor Receptor – Role of Heparin in the Activation of the Receptor. Biochemistry 44(48):15787-98. 2005. (COBRE funding acknowledged).
- RAJALINGAM, DAKSHINAMURTHY.; KUMAR, SURESH T. K.; YU, CHIN. C2A Domain of Synaptotagmin Exhibits High Binding Affinity for Copper – Implications in the Formation of the Multiprotein Release Complex. Biochemistry 44(44):14431-42. 2005 (COBRE funding acknowledged).
- SIVARAJA, VAITHIYALINGAM.; KUMAR, SURESH T. K.; PRUDOVSKY, IGOR.; YU, CHIN. Three-Dimensional Solution Structure of a Unique S100 Protein. Biochem. Biophys. Res. Commun., 335(4):1140-8. 2005 (COBRE funding acknowledged).
- HSIEH, HUI-CHU.; KUMAR, SURESH T. K.; CHIU, CHI-CHENG.; YU, CHIN. Equilibrium Unfolding of an Oligomeric Protein Involves the Formation of a Multimeric Intermediate. Biochem. Biophys. Res. Commun. 326(1):108-14. 2005. (COBRE funding acknowledged).
- KATHIR, KARUPPANAN.; KUMAR, SURESH T. K.; RAJALINGAM, DAKSHINAMURTHY.; YU, CHIN. Time-dependent Changes in the Denatured State(s) Affect Folding Mechanism of an All Beta-Sheet Protein. J. Biol. Chem. 280(33):29682-8. 2005 (COBRE funding acknowledged).
- SIVARAJA, VAITHIYALINGAM.; KUMAR, SURESH T. K.; YU, CHIN. Resonance Assignments for Mouse S100A13 Assignment of 1H, 13C, and 15N Resonances in S100A13 J. Biomol. NMR, 32(3):257-58. 2005 (COBRE funding acknowledged
- JAYACHITRA, KANDASAMY.; KUMAR, SURESH T. K.; LU, TIAN-JU.; YU, CHIN.; CHIN, DER-HANG. Cold Instability of Aponeocarzinostatin and its stabilization by Labile Chromophore Biophys. J. 88(6):4252-61. 2005 (COBRE funding acknowledged)
- HSIEH, HUI-CHU.; SURESH, KUMAR T. K.; SIVARAMAN,THIRUNAVKARASU.; YU, CHIN. Refolding of a Small All Beta-Sheet Protein Proceeds with Accumulation of Kinetic Intermediates Arch. Biochem. Biophys. (in press) 2006 (COBRE funding acknowledged).