Project 5
Molecular basis of the aberrant signaling behavior of Ras-related proteins that lead to cancer
(Paul Adams)
A major challenge still facing the biomedical community is the limited development of therapeutic strategies to tackle the oncogenic potential of the Ras super-family of proteins. Strategies that will have high impact on future drug design must target new molecular features of cell signaling proteins involved in cell proliferation and transformation, as well as other oncogenic events. The activity of Ras-related proteins, is dictated by its nucleotide-bound state (GTP-bound active; GDP-bound form inactive). These states can be distinguished by conformational differences, most of which are localized in two relatively flexible “Switch” regions as characterized by internal dynamics on multiple timescales, and this flexibility may be essential for protein interactions, some of which can lead to abnormal cell-signaling activity involving Ras-related proteins.
Our long-term research goals are to understand the molecular basis of the aberrant signaling behavior of Ras-related proteins that lead to cancer, facilitating approaches to the identification and development of potential targets for therapeutic design. Presently, we are using molecular biology, as well as, biophysical and biochemical techniques in the design of strategies to study molecular details of the Ras proteins Cdc42 and Rheb that we expect to aid our understanding of how changes in conformation/dynamics of these proteins and interaction with regulator/effectors proteins affect various important Ras-regulated signal transduction pathways.
Recently, studies from our laboratory on the biomolecular dynamics, structure, and biological function of a mutant of Cdc42, relative to wild type protein highlighted that, in addition to strategies to block Ras-protein interactions, strategies to restrict the conformational flexibility of important binding regions of Ras leading to the inhibition of effector interactions that facilitate aberrant signaling activity may also have therapeutic potential (Chandreshekar, et al., 2011, Biochemistry, 50, p. 6196-207).
With a better understanding of conformational changes in Ras proteins that affect normal cell-signaling, we expect to provide information at the molecular level that will shed light on mechanisms of specific signal transduction pathways regulating oncogenic events.
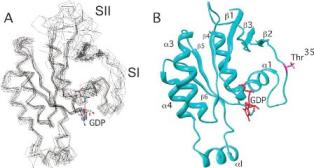
Structure of Cdc42(T35A)-GDP, a single-point mutant construct of the Ras protein Cdc42 in a region known to be important in effector protein interactions.
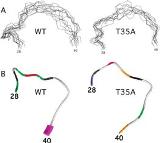
(A) Stereoview of the backbone ensemble of the Switch I region of wild-type Cdc42 and Cdc42(T35A). (B) Backbone of the Switch I region colored on the basis of best-fit dynamics model assignments from dynamics calculations. From reference (1).
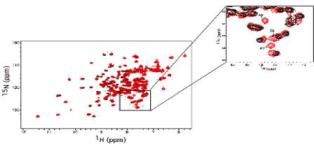
2D 15N-HSQC of Cdc42 wildtype (black) overlaid with Cdc42T35A (red). The inset (upper right corner of figure) highlights a region of the spectrum containing resonances from three residues on the C-terminal portion of the Switch 1 region of Cdc42T35A (Residues 38, 40 and 41) not detected in the HSQC spectrum of Cdc42 wild type. from reference (1)
Recent Publications
R. Chandrashekar, O. Salem, H. Krizova, R. McFeeters and P. D. Adams, “A switch 1 mutant of Ras protein Cdc42 exhibits decreased conformational freedom”, 2011, Biochemistry, 50(28):6196-207.
K. Kathir, L. Gao, D. Rajalingam, A. Daily, S. Brixey, H. Liu, D. Davis, P.D. Adams, I. Prudovsky, and T.K.S. Kumar, “NMR Characterization of the Copper and Lipid Interactions of the C2B Domain of Synaptotagmin I-Relevance to the Non Classical Secretion of the Human Acidic Fibroblast Growth Factor (hFGF-1)”, Biochim. Biophys. Acta, (2010), vol 1798, #2, p. 297-302.
D. Rajalingam, K. Kathir, K. Ananthamurthy, P.D. Adams, T.K.S. Kumar, “An Efficient Method to Prevent the Degradation of Recombinant Proteins by Thrombin,” Analytical Biochemistry, (2008), 375, p.361-363
R.E. Oswald and P.D. Adams, “NMR Assignment of Cdc42(T35A), an Active Switch I Mutant of Cdc42,” Biomolecular NMR Assignments, (2007), 1: p. 225-227.