Project 6
Physiology, Biochemistry, and Genetics of Anaerobic Methane-producing Methanogens
(Dr. Daniel Lessner)
My research program is focused on the physiology, biochemistry, and genetics of the strictly anaerobic methane-producing archaea (methanogens). Methanogens are found in virtually every anaerobic environment and are the only organisms capable of biological methane production. Methane produced by methanogens is critical to the global cycling of carbon, a potent greenhouse gas, and a valuable biofuel. The pathways and enzymes involved in methanogenesis have been identified and well-characterized. However, much less is known about how methanogens respond to environmental change, including stressors.
My research program is focused on the physiology, biochemistry, and genetics of the strictly anaerobic methane-producing archaea (methanogens). Methanogens are found in virtually every anaerobic environment and are the only organisms capable of biological methane production. Methane produced by methanogens is critical to the global cycling of carbon, a potent greenhouse gas, and a valuable biofuel. The pathways and enzymes involved in methanogenesis have been identified and well-characterized. However, much less is known about how methanogens respond to environmental change, including stressors.
Because many methanogenesis enzymes contain oxygen-labile cofactors such as iron-sulfur clusters (1), we are particularly interested in understanding how methanogens sense and respond to oxygen and oxidative stress. We are using Methanosarcina acetivorans as a model organism, due to its metabolic diversity and robust genetic system. Two primary projects are focused on M. acetivorans proteins that contain regulatory iron-sulfur clusters. In each protein, the iron-sulfur clusters are proposed to function by modulating the structure and activity of the protein in response to oxygen or reactive oxygen species. The results obtained will provide insight into the function of iron-sulfur clusters in proteins involved in response to oxidative stress. Overall, an understanding of the oxidative stress response in methanogens will lead to better methods for modulating biological methane production, as well as providing insight into how anaerobes and archaea in general cope with oxidative stress.
A third project is focused on utilizing the established M. acetivorans genetic system to develop methanogen strains with improved capabilities, including increased substrate utilization and oxygen tolerance.
Project 1: Unraveling the role of iron-sulfur clusters in RNA polymerase
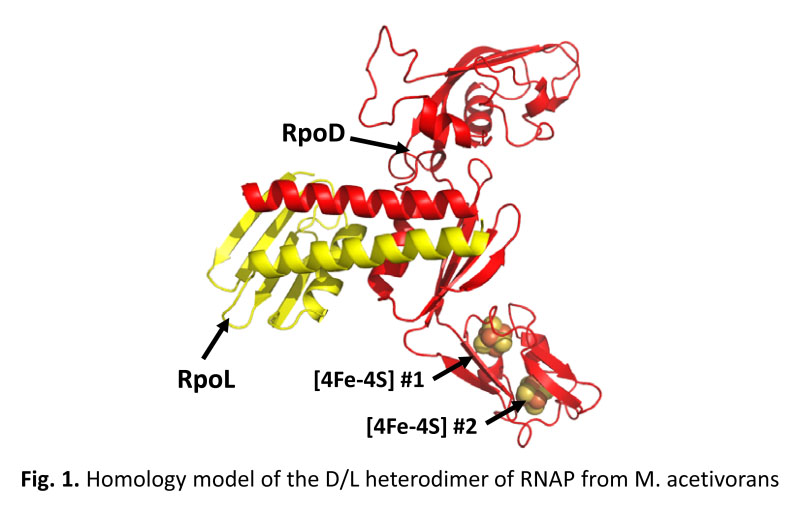
Recently, the RpoD subunit of RNA polymerase (RNAP) in several archaea, including methanogens, and the corresponding Rpb3 subunit of RNAP in several eukaryotes were shown or predicted to contain a [4Fe-4S] cluster. The function of the [4Fe-4S] cluster is unknown. RpoD forms a heterodimer with RpoL, which is the first step in assembly of multi-subunit RNAP in Archaea. The clusters are postulated to regulate the interaction of RpoD with RpoL or other subunits to control RNAP assembly and activity in response to oxidants. M. acetivorans RpoD is predicted to bind two [4Fe-4S] clusters within a domain that is likely not required for interaction with RpoL (Fig. 1). We are using a combination of in vivo mutational analyses and native and recombinant RpoDL/RNAP purification to elucidate the importance of the clusters to RNAP assembly and activity (4). This project is currently funded by the NSF.
Project 2: Understanding the role of metal cofactors as molecular switches in the MdrA family of disulfide reductases
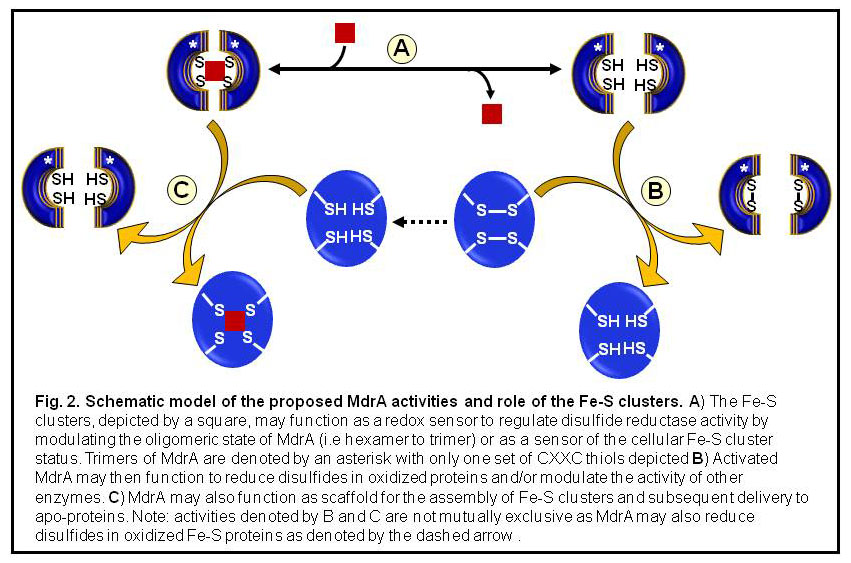
We have identified a novel iron-sulfur cluster containing disulfide reductase (MdrA) from M. acetivorans (2).The iron-sulfur clusters in MdrA are postulated to function as a molecular switch to control disulfide reductase activity. MdrA may function to repair of oxidatively-damaged iron-sulfur clusters (Fig. 2). We are using a combination of genetics and biochemistry to determine the precise physiological role(s) of MdrA. Through collaboration with Dr. Suresh Kumar (Dept. of Chemistry & Biochemistry) we are using biophysical techniques (NMR, CD, ITC, and DSC) to examine the influence of the clusters, and other metals, on the structure and properties of MdrA. This project is currently supported by the Arkansas Biosciences Institute and the NIH COBRE center.
Project 3: Genetic engineering of methanogens
We are generating strains of M. acetivorans with improved characteristics, using genetic
engineering and directed evolution approaches. We have recently produced a strain
of M. acetivorans with a hybrid methanogenesis pathway that confers the ability to
grow and produce methane with non-native substrates (Fig. 3) (3). We are currently
constructing strains with increased oxygen tolerance and metabolic capabilities
Publications
1. Lessner, D. 2009. Methanogenesis: Biochemistry, Encyclopedia of Life Sciences. Nature Publishing Group.
2. Lessner, D. J., and J. G. Ferry. 2007. The Archaeon Methanosarcina acetivorans Contains a Protein Disulfide Reductase with an Iron-Sulfur Cluster. J Bacteriol 189:7475-84.
3. Lessner, D. J., L. Lhu, C. S. Wahal, and J. G. Ferry. 2010. An engineered methanogenic pathway derived from the domains Bacteria and Archaea. mBio 1:e000243-10.
4. Lessner, F. H., M. E. Jennings, A. Hirata, E. C. Duin, and D. J. Lessner. 2011. Subunit D of RNA polymerase from Methanosarcina acetivorans contains two oxygen-labile [4Fe-4S] clusters: implications for regulation of transcription in archaea in preparation.